In our latest journal club, we discussed the biosynthesis of tetrahymanol (Fig. 1) in Bacteria 1, which is of importance in organic geochemistry as it is the precursor of the biomarker gammacerane.

Gammacerane is a pentacyclic triterpenoid (Fig. 1) and was documented for the first time in the Green River Shale in 1966 2, while its oldest documented occurrence was in the Late Paleoproterozoic 3, 850 million years ago. For a while, it was not really clear where gammacerane came from, but its presence in a lot of halites and evaporites, rocks formed by evaporation of seawater, lead to the assumption that it was associated with hypersaline environments 4,5. It is present in many oils as well, and the gammacerane index6 is widely used to describe and identify oils and source rocks.
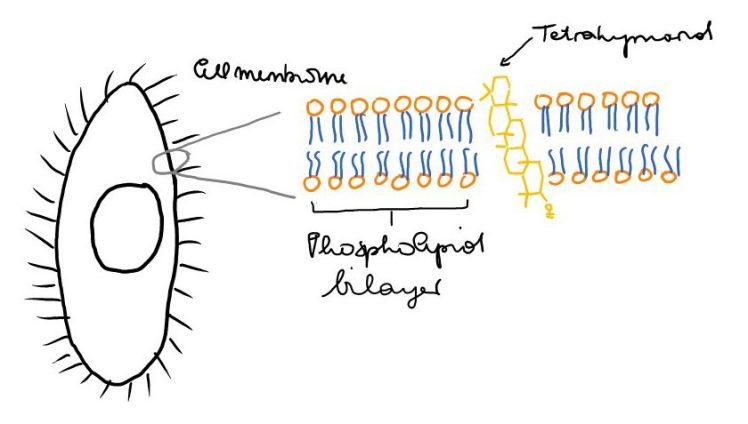
In the late 80s/early 90s, it became clear that the biological precursor of gammacerane is tetrahymanol (Fig. 2), a pentacyclic triterpenoidal alchohol 7. Tetrahymanol was, for the first time, isolated from a eukaryote, the ciliate Tetrahymena pyriformis by Mallory et al. 8 and its structure further confirmed by synthesis and crystallisation 9,10. It was also found to be present in T. thermophila 11, and widespread in marine sediments where tetrahymanol-producing ciliates were abundant in the overlying water column 12–14. Its function in the membranes of these organisms seems to be similar to the function of sterols15. These ciliates are actually deficient in the genes for sterol synthesis 16 – and mostly, they cope with this sterol deficiency by taking up sterols and incorporating them into their membranes. However, they possess an enzyme that can catalyse the cyclisation of squalene to tetrahymanol 20, and when no sterols are available, the enzyme that makes tetrahymanol kicks in! However, when sterols such as ergosterol or cholesterol are provided, the enzyme that makes tetrahymanol is inhibited 17–19. Most importantly, tetrahymanol synthesis does not require molecular oxygen, contrary to sterol synthesis 21,22 – the capability of producing it thus enables ciliates to thrive in anaerobic environments, where they are less likely to find a supply of sterols from – oxygen requiring – sterol producing organisms. Sinninghe Damsté et al. 23 recognised this and showed that the carbon isotopic composition of gammacerane was similar to the one of a biomarker of anaerobic bacteria, which is what they suggest that ciliates must have been feeding on instead of sterol-producing organisms (bacteria don’t usually produce sterols). Gammacerane was thus defined as a biomarker for a stratified water column, typical for settings with photic zone euxinia, and generally co-occurs with biomarkers that fit into these settings as well.
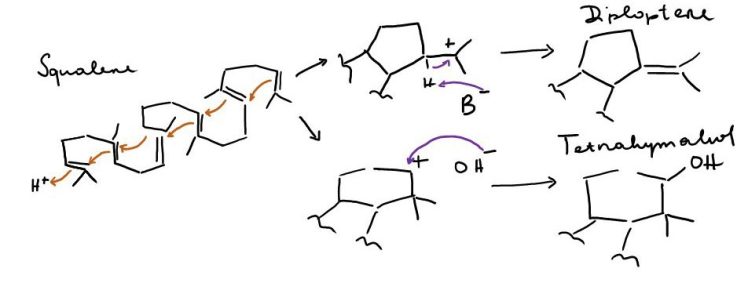
Tetrahymanol in ciliates is produced from squalene by squalene-tetrahymanol cyclase, stc 20,24,25 – these ciliates lack both the enzymes for sterol synthesis usually present in eukaryotes, as well as the enzymes for hopane synthesis usually present in bacteria (squalene-hopane-cyclase shc) 16. Stc can catalyse both the cyclisation of squalene into a four-ring system plus a 5-ring and a double bond via a tertiary carbocation, the more stable intermediate, but also into a four-ring system plus a 6-ring with a hydroxylation (Fig. 3). Interestingly, this pathway doesn’t seem to be unique to ciliates – also the fern Oleandra wallichii 26 and the rumen fungus Piromonas communis 27 possess stc and have been shown to produce tetrahymanol. Takishita et al. 28 have recently shown that also a few select other protists (other ciliates, flagellates), a polychaete, and another fungus also possess this gene and at least some of these organisms have also been shown to produce tetrahymanol. According to their analysis, all the stc sequences are closely related and must have been transferred laterally.
However, of equally great interest is the presence of tetrahymanol in different Bacteria: Rhodopseudomonas palustris 29 and Bradyrhizobium japonicum 30. While B. japonicum is a nitrogen-fixing root-symbiont, R. palustris is a metabolically rather versatile bacterium that has been found in many environments, including the marine environment. R. palustris biomass can consist to 0.4 mg/g of tetrahymanol 29! However, the biosynthesis of tetrahymanol in those bacteria is less clear, as they do not possess stc, but squalene-hopane cyclase shc 31,32, which is very widespread in many bacteria, but produces a different compound (four-ring system plus a 5-ring) 33–36. It was hypothesised that the production of tetrahymanol by shc, which usually catalyses the cyclisation to a different hopane (diploptene) is due to a high versatility of shc 35, but the purified enzyme from those two bacteria could not produce tetrahymanol from squalene 31,32.
Tetrahymanol biosynthesis in bacteria
There are various reasons why determining the biosynthetic pathway for tetrahymanol synthesis in bacteria is of great interest. First of all, knowledge about genes involved in the production of tetrahymanol would give a better understanding of evolutionary development. But secondly, and most importantly for Organic Geochemistry, the knowledge of the specific gene catalysing tetrahymanol synthesis will allow data mining of genomic databases and environmental metagenomes and transcriptomes. This will give us a better overview over the various tetrahymanol producers in addition to ciliates, and allow to further investigate the links with hypersaline and anaerobic, stratified depositional settings. In a recently published article in PNAS, Banta et al.1 attempt to identify specific biosynthesis genes responsible for tetrahymanol synthesis in bacteria and investigate whether particular environmental conditions regulate the production of tetrahymanol in bacteria.
At the first stages of their investigations, they observed the production of tetrahymanol in Methylomicrobium alcaliphilum (γ-Proteobacteria). It was the first time that tetrahymanol was observed in γ-Proteobacteria and, surprisingly, M. alcaliphilum produced significantly more tetrahymanol than what was typically observed in α-Proteobacterial species. From the article, it is not clear what the motivation for testing M. alcaliphilum was, what other bacteria they had tested at this stage or what the conditions for culturing were. Then, to identify putative bacterial tetrahymanol biosynthesis proteins and the protein-encoding genes, they compared the genomic make-up, they found protein-encoding genes present in M. alcaliphilum, and the well-known tetrahymanol-producers, R. palustris and B. japonicum, yet absent in non-tetrahymanol producing bacteria. Eventually, they found one gene, encoding a hypothetical protein with no identifiable motifs: MEALZ_1626.
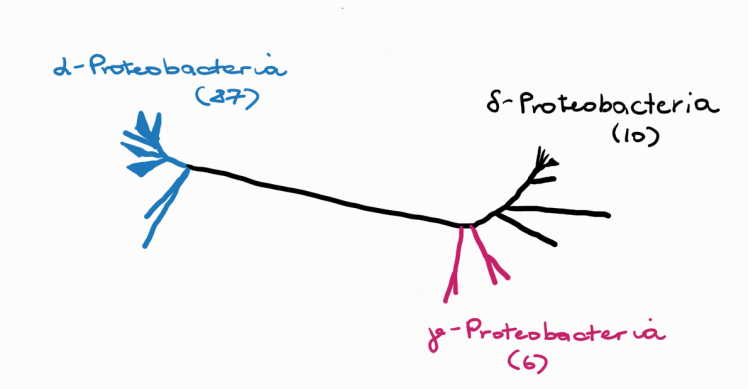
When testing it experimentally by knock-out experiments, they could confirm that MEALZ_1626 indeed encodes a protein that is capable of synthesizing tetrahymanol with squalene hopane cyclase – Tetrahymanol synthase, Ths. Mining of genomic databases confirmed the presence of Ths in 104 bacterial genomes in three different phyla: α-Proteobacteria, aerobic methanotrophic γ-Proteobacteria and also sulfate-reducing δ-Proteobacteria. Therefore, a very significant finding of this project is that the capacity for tetrahymanol biosynthesis is more widespread in the bacterial domain than previously thought.
However, there are actually two divergent lineages of Ths:
- α-Proteobacteria, which covers the majority of the tetrahymanol producing bacteria
- γ-Proteobacterial methanotrophs and δ-Proteobacterial sulfate- reducers, which are the minority in the studied bacteria (Fig. 4)
Apart from the types of tetrahymanol producing bacteria; an equally significant aspect for the paleoenvironmental study is the linkage between biosynthesis of a specific molecule and a specific type of ecosystem, environment and climate. To make it possible, Banta and co-authors took a step forward towards environmental metagenome studies. When looking for homologs of Ths in environmental metagenomes, 472 potential Ths homologs were identified in 35 different metagenomes (restricted to freshwater, soil, and marine environments), including 25 soil, 8 freshwater, and 2 marine metagenomes. This observation indicates terrestrial or lacustrine ecosystems as a significant source of bacterial tetrahymanol. However, marine ciliates are still likely to be the predominant source in marine environments – though the authors do not present details on how much marine data were analysed for Ths.
In addition, since the production of tetrahymanol in aerobic methanotrophs and sulfate-reducing bacteria is restricted to a few types of bacteria, Banta and co-authors propose a common environment triggering production of tetrahymanol in bacteria and ciliates. For instance, it can reflect stratified environments, as the occurrence of aerobic methanotrophs in the suboxic zone of stratified marine and freshwater water bodies, and sulfate-reducing bacteria in the anoxic sediments of these systems have been documented.
Interestingly, the genes for the biosynthesis of tetrahymanol in bacteria are not related at all to the genes in eukaryotes – a phenomenon called convergent evolution, distinct mechanisms to synthesise the exact same molecule (tetrahymanol) were developed. This could indicate the significance of tetrahymanol structure and might be a key toward finding its specific functionality.
This paper presents a significant step forward in the understanding of tetrahymanol biosynthesis in bacteria and answers a long-standing puzzle. However, the big question still remains: Why and under what type of conditions does the bacterial community prefer to synthesise tetrahymanol rather than hopanoids? And how will this influence interpretations of the gammacerane biomarker record?
References
- Banta, A. B., Wei, J. H. & Welander, P. V. A distinct pathway for tetrahymanol synthesis in bacteria. Proc. Natl. Acad. Sci. 112, 13478–13483 (2015).
- Hills, I. R., Whitehead, E. V., Anders, D. E., Cummins, J. J. & Robinson, W. E. An optically active triterpane, gammacerane in Green River, Colorado, oil shale bitumen. Chem. Commun. Lond. 752b (1966). doi:10.1039/c1966000752b
- Summons, R. E. et al. Distinctive hydrocarbon biomarkers from fossiliferous sediment of the Late Proterozoic Walcott Member, Chuar Group, Grand Canyon, Arizona. Geochim. Cosmochim. Acta 52, 2625–2637 (1988).
- Kenig, F., Sinninghe Damsté, J. S., Frewin, N. L., Hayes, J. M. & De Leeuw, J. W. Molecular indicators for palaeoenvironmental change in a Messinian evaporitic sequence (Vena del Gesso, Italy). II: High-resolution variations in abundances and 13C contents of free and sulphur-bound carbon skeletons in a single marl bed. Org. Geochem. 23, 485–526 (1995).
- Moldowan, J. M., Seifert, W. K. & Gallegos, E. J. Relationship Between Petroleum Composition and Depositional Environment of Petroleum Source Rocks. AAPG Bull. 69, 1255–1268 (1985).
- Peters, K. E., Walters, C. C. & Moldowan, J. M. The Biomarker Guide. (Cambridge University Press, 2004).
- Haven, H. L. T., Rohmer, M., Rullkötter, J. & Bisseret, P. Tetrahymanol, the most likely precursor of gammacerane, occurs ubiquitously in marine sediments. Geochim. Cosmochim. Acta 53, 3073–3079 (1989).
- Mallory, F. B., Gordon, J. T. & Conner, R. L. The Isolation of a Pentacyclic Triterpenoid Alcohol from a Protozoan. J. Am. Chem. Soc. 85, 1362–1363 (1963).
- Langs, D. A., Duax, W. L., Carrell, H. L., Berman, H. & Caspi, E. Crystal structure of tetrahymanol hemihydrate. J. Org. Chem. 42, 2134–2137 (1977).
- Tsuda, Y. et al. The synthesis of tetrahymanol. Tetrahedron Lett. 6, 1427–1431 (1965).
- Holz, G. G. & Conner, R. L. in Biology of Tetrahymena (ed. Elliott, A. M.) 99–122 (Dowden, Hutchinson & Ross, 1973).
- Harvey, H. R. & Mcmanus, G. B. Marine ciliates as a widespread source of tetrahymanol and hopan-3β-ol in sediments. Geochim. Cosmochim. Acta 55, 3387–3390 (1991).
- Venkatesan, M. I. Tetrahymanol: Its widespread occurrence and geochemical significance. Geochim. Cosmochim. Acta 53, 3095–3101 (1989).
- Venkatesan, M. I., Ruth, E. & Kaplan, I. R. Triterpenols from sediments of Santa Monica Basin, Southern California Bight, U.S.A. Org. Geochem. 16, 1015–1024 (1990).
- Nozawa, Y., Fukushima, H. & Iida, H. Studies on Tetrahymena membranes. Biochim. Biophys. Acta BBA – Biomembr. 406, 248–263 (1975).
- Tomazic, M. L., Poklepovich, T. J., Nudel, C. B. & Nusblat, A. D. Incomplete sterols and hopanoids pathways in ciliates: Gene loss and acquisition during evolution as a source of biosynthetic genes. Mol. Phylogenet. Evol. 74, 122–134 (2014).
- Conner, R. L. et al. Ergosterol replacement of tetrahymanol in Tetrahymena membranes. Biochem. Biophys. Res. Commun. 44, 995–1000 (1971).
- Raederstorff, D. & Rohmer, M. Polyterpenoids as cholesterol and tetrahymanol surrogates in the ciliate Tetrahymena pyriformis. Biochim. Biophys. Acta BBA – Lipids Lipid Metab. 960, 190–199 (1988).
- Warburg, C. F., Wakeel, M. & Wilton, D. C. The role of squalene synthetase in the inhibition of tetrahymanol biosynthesis by cholesterol inTetrahymena pyriformis. Lipids 17, 230–234 (1982).
- Caspi, E. Biosynthesis of tetrahymanol by Tetrahymena pyriformis: mechanistic and evolutionary implications. Acc. Chem. Res. 13, 97–104 (1980).
- Summons, R. E., Bradley, A. S., Jahnke, L. L. & Waldbauer, J. R. Steroids, triterpenoids and molecular oxygen. Philos. Trans. R. Soc. B Biol. Sci. 361, 951–968 (2006).
- Waldbauer, J. R., Newman, D. K. & Summons, R. E. Microaerobic steroid biosynthesis and the molecular fossil record of Archean life. Proc. Natl. Acad. Sci. 108, 13409–13414 (2011).
- Sinninghe Damsté, J. S. et al. Evidence for gammacerane as an indicator of water column stratification. Geochim. Cosmochim. Acta 59, 1895–1900 (1995).
- Saar, J., Kader, J.-C., Poralla, K. & Ourisson, G. Purification and some properties of the squalene-tetrahymanol cyclase from Tetrahymena thermophila. Biochim. Biophys. Acta BBA – Gen. Subj. 1075, 93–101 (1991).
- Bouvier, P., Berger, Y., Rohmer, M. & Ourisson, G. Non-specific Biosynthesis of Gammacerane Derivatives by a Cell-Free System from the Protozoon Tetrahymena pyriformis: Conformations of Squalene, (3S)-Squalene Epoxide and (3R)-Squalene Epoxide during the Cyclization. Eur. J. Biochem. 112, 549–556 (2005).
- Zander, J. M., Caspi, E., Pandey, G. N. & Mitra, C. R. The presence of tetrahymanol in Oleandra wallichii. Phytochemistry 8, 2265–2267 (1969).
- Kemp, P., Lander, D. J. & Orpin, C. G. The Lipids of the Rumen Fungus Piromonas communis. Microbiology 130, 27–37 (1984).
- Takishita, K. et al. Lateral transfer of tetrahymanol-synthesizing genes has allowed multiple diverse eukaryote lineages to independently adapt to environments without oxygen. Biol. Direct 7, 5 (2012).
- Kleemann, G. et al. Tetrahymanol from the phototrophic bacterium Rhodopseudomonas palustris: first report of a gammacerane triterpene from a prokaryote. J. Gen. Microbiol. 136, 2551–2553 (1990).
- Bravo, J.-M., Perzl, M., Härtner, T., Kannenberg, E. L. & Rohmer, M. Novel methylated triterpenoids of the gammacerane series from the nitrogen-fixing bacterium Bradyrhizobium japonicum USDA 110: Triterpenoids of Bradyrhizobium japonicum. Eur. J. Biochem. 268, 1323–1331 (2001).
- Kleemann, G., Kellner, R. & Poralla, K. Purification and properties of the squalene-hopene cyclase from Rhodopseudomonas palustris, a purple non-sulfur bacterium producing hopanoids and tetrahymanol. Biochim. Biophys. Acta BBA – Lipids Lipid Metab. 1210, 317–320 (1994).
- Perzl, M., Muller, P., Poralla, K. & Kannenberg, E. L. Squalene-hopene cyclase from Bradyrhizobium japonicum: cloning, expression, sequence analysis and comparison to other triterpenoid cyclases. Microbiology 143, 1235–1242 (1997).
- Pearson, A. in Treatise on Geochemistry 291–336 (Elsevier, 2014).
- Fischer, W. W. & Pearson, A. Hypotheses for the origin and early evolution of triterpenoid cyclases. Geobiology 0, 070210031741002–??? (2007).
- Siedenburg, G. & Jendrossek, D. Squalene-Hopene Cyclases. Appl. Environ. Microbiol. 77, 3905–3915 (2011).
- Abe, I. Enzymatic synthesis of cyclic triterpenes. Nat. Prod. Rep. 24, 1311 (2007).